TORONTO, May 28: Dozens of genes, previously thought to have similar roles across different organisms, are in fact unique to humans and could help explain how our species evolved, scientists have found.
These genes code for a class of proteins known as transcription factors, or TFs, which control gene activity, according to researchers at the University of Toronto in Canada.
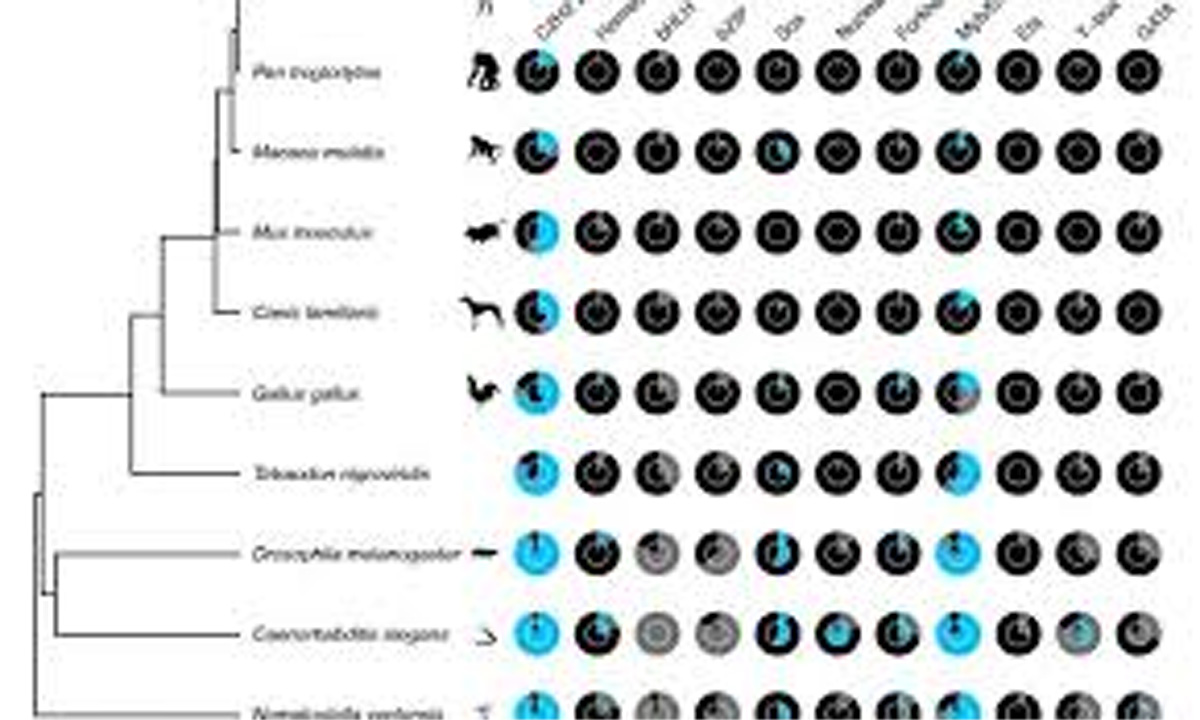
TFs recognise specific snippets of the DNA code called motifs, and use them as landing sites to bind the DNA and turn genes on or off.
Previous research had suggested that TFs which look similar across different organisms also bind similar motifs, even in species as diverse as fruit flies and humans.
However, a new study from Professor Timothy Hughes’ lab, from the University of Toronto, shows that this is not always the case.
The researchers found a new computational method which allowed them to more accurately predict motif sequences each TF binds in many different species.
The findings, published in the journal Nature Genetics, reveal that some sub-classes of TFs are much more functionally diverse than previously thought.
“Even between closely related species there’s a non-negligible portion of TFs that are likely to bind new sequences,” said Sam Lambert, former graduate student in Hughes’ lab.
“This means they are likely to have novel functions by regulating different genes, which may be important for species differences,” said Lambert, who is now at the University of Cambridge in the UK.
Even between chimps and humans, whose genomes are 99 per cent identical, there are dozens of TFs which recognise diverse motifs between the two species in a way that would affect expression of hundreds of different genes.
“We think these molecular differences could be driving some of the differences between chimps and humans,” said Lambert.
To reanalyse motif sequences, Lambert developed new software which looks for structural similarities between the TFs’ DNA binding regions that relate to their ability to bind the same or different DNA motifs.
If two TFs, from different species, have a similar composition of amino-acids, building blocks of proteins, they probably bind similar motifs.
However, unlike older methods, which compare these regions as a whole, the new method automatically assigns greater value to those amino-acids — a fraction of the entire region — which directly contact the DNA.
In this case, two TFs may look similar overall, but if they differ in the position of these key amino-acids, they are more likely to bind different motifs.
When Lambert compared all TFs across different species and matched to all available motif sequence data, he found that many human TFs recognise different sequences — and therefore regulate different genes — than versions of the same proteins in other animals.
The finding contradicts earlier research, which stated that almost all of human and fruit fly TFs bind the same motif sequences, and is a call for caution to scientists hoping to draw insights about human TFs by only studying their counterparts in simpler organisms.
“There is this idea that has persevered, which is that the TFs bind almost identical motifs between humans and fruit flies,” said Hughes.
“And while there are many examples where these proteins are functionally conserved, this is by no means to the extent that has been accepted,” he said.
As for TFs that have unique human roles, these belong to the rapidly evolving class of so-called C2H2 zinc finger TFs, named for zinc ion-containing finger-like protrusions, with which they bind the DNA.
Their role remains an open question but it is known that organisms with more diverse TFs also have more cell types, which can come together in novel ways to build more complicated bodies.
Hughes is excited about a tantalising possibility that some of these zinc finger TFs could be responsible for the unique features of human physiology and anatomy — our immune system and the brain, which are the most complex among animals. (PTI)